Table of Contents
Estimate univariate and multivariate sample skewness and kurtosis in popular statistical software
This tutorial explains four different methods for calculating univariate and multivariate skewness and kurtosis in your data: an online calculator that allows the use of Excel, SPSS, and SAS data sets directly as well as text and csv files, an SPSS program, a SAS program, and instructions for STATA.
Online calculator
This introduces the online tool to calculate univariate/multivariate skewness and kurtosis at http://webpower.psychstat.org/models/kurtosis/
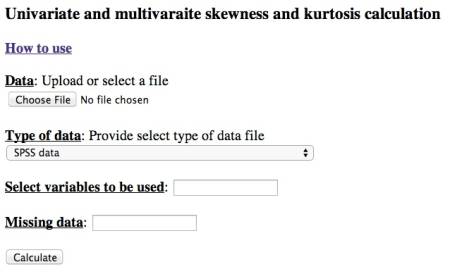
The interface of the tool looks like
Data file
The data file can be chosen by clicking the Choose File button (it might appear differently for different browsers). We DO NOT save your data file and it is deleted immediately after calculation.
Type of data
The following types of data are allowed:
- SPSS data file with the extension name .sav
- SAS data file with the extension name .sas7bdat
- Excel data file with the extension name .xls or .xlsx
- CSV file (comma separated value data file) with extension name .csv
- TXT file (text file) with extension name .txt
Select variables
A subset of variables can be used. To use the whole data set, leave this field blank. To select a subset of variables, provide the column numbers that separated by comma (,). For example
1, 2-5, 7-9, 11
will select variables 1, 2, 3, 4, 5, 7, 8, 9, 11
Missing data
Missing data values can be provided. If multiple values are used to denote missing data, they can be separated by comma (,). For example,
-999, -888, NA
will replace all three values above to missing data.
R
R functions for the calculation
library(MASS)
skewkurt<-function(x, na.rm=TRUE){
if (na.rm) x <- x[!is.na(x)]
n <- length(x)
skew1 <- (sum((x - mean(x))^3)/n)/(sum((x - mean(x))^2)/n)^(3/2)
skew2 <- n/((n-1)*(n-2))*sum((x - mean(x))^3)/(sd(x))^3
skew.se <- sqrt(6*n*(n-1) / ((n-2)*(n+1)*(n+3)))
temp<- sum((x - mean(x))^4)/(sum((x - mean(x))^2)^2)
kurt1 <- n * temp -3
kurt2 <- n*(n+1)*(n-1)/((n-2)*(n-3))*temp - 3*(n-1)^2/((n-2)*(n-3))
kurt.se <- sqrt(4*(n^2-1)*skew.se^2 / ((n-3)*(n+5)))
list(skew1=skew1, skew2=skew2, skew.se=skew.se, kurt1=kurt1, kurt2=kurt2, kurt.se=kurt.se)
}
multi.skewkurt<-function (x, na.rm = TRUE){
x <- as.matrix(x)
if (na.rm)
x <- na.omit(x)
n <- dim(x)[1]
p <- dim(x)[2]
x <- scale(x, scale = FALSE)
S <- cov(x)*(n-1)/n
#S.inv <- solve(S)
S.inv <- ginv(S)
D <- x %*% S.inv %*% t(x)
b1p <- sum(D^3)/n^2
b2p <- tr(D^2)/n
chi.df <- p * (p + 1) * (p + 2)/6
k <- (p + 1) * (n + 1) * (n + 3)/(n * ((n + 1) * (p + 1) -
6))
small.skew <- n * k * b1p/6
M.skew <- n * b1p/6
M.kurt <- (b2p - p * (p + 2)) * sqrt(n/(8 * p * (p + 2)))
p.skew <- 1 - pchisq(M.skew, chi.df)
p.small <- 1 - pchisq(small.skew, chi.df)
p.kurt <- 2 * (1 - pnorm(abs(M.kurt)))
results <- list(n.obs = n, n.var = p, b1p = b1p, b2p = b2p,
skew = M.skew, small.skew = small.skew, p.skew = p.skew,
p.small = p.small, kurtosis = M.kurt, p.kurt = p.kurt)
return(results)
}
mardia<-function(x, na.rm=TRUE){
if (na.rm)
x <- na.omit(x)
n <- dim(x)[1]
p <- dim(x)[2]
uni <- function(x){
n <- length(x)
xbar <- mean(x)
m2 <- sum((x-xbar)^2)/n
m3 <- sum((x-xbar)^3)/n
m4 <- sum((x-xbar)^4)/n
skewness <- sqrt(n*(n-1))/(n-2)*m3/m2^1.5
kurtosis <- (n-1)/((n-2)*(n-3))*((n+1)*(m4/m2^2-3)+6)
skew.se <- sqrt(6*n*(n-1) / ((n-2)*(n+1)*(n+3)))
kurt.se <- sqrt(4*(n^2-1)*skew.se^2 / ((n-3)*(n+5)))
c(skewness, skew.se=skew.se, kurtosis, kurt.se)
}
univariate <- apply(x, 2, uni)
rownames(univariate) <- c('Skewness', 'SE_skew', 'Kurtosis', 'SE_kurt')
x <- scale(x, scale = FALSE)
S <- cov(x)*(n-1)/n
S.inv <- ginv(S)
D <- x %*% S.inv %*% t(x)
b1p <- sum(D^3)/n^2
b2p <- sum(diag(D^2))/n
chi.df <- p * (p + 1) * (p + 2)/6
k <- (p + 1) * (n + 1) * (n + 3)/(n * ((n + 1) * (p + 1) - 6))
small.skew <- n * k * b1p/6
M.skew <- n * b1p/6
M.kurt <- (b2p - p * (p + 2)) * sqrt(n/(8 * p * (p + 2)))
p.skew <- 1 - pchisq(M.skew, chi.df)
p.small <- 1 - pchisq(small.skew, chi.df)
p.kurt <- 2 * (1 - pnorm(abs(M.kurt)))
multivariate <- rbind(c(b1p, M.skew, p.skew), c(b2p, M.kurt, p.kurt))
rownames(multivariate) <- c('Skewness', 'Kurtosis')
colnames(multivariate) <- c('b', 'z', 'p-value')
results <- list(n.obs = n, n.var = p, univariate = univariate, multivariate=multivariate)
cat('Sample size: ', n, "\n")
cat('Number of variables: ', p, "\n\n")
cat("Univariate skewness and kurtosis\n")
print(t(univariate))
cat("\nMardia's multivariate skewness and kurtosis\n")
print(multivariate)
cat("\n")
invisible(results)
}
SPSS
An SPSS macro developed by Dr. Lawrence T. DeCarlo needs to be used. We have edited this macro to get the skewness and kurtosis only.
First, download the macro (right click here to download) to your computer under a folder such as c:\Users\johnny\.
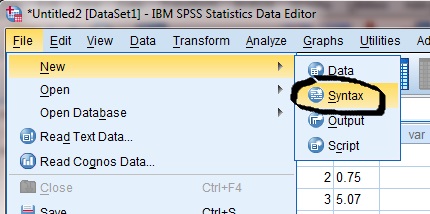
Second, open a script editor within SPSS
Third, in the script editor, type the following
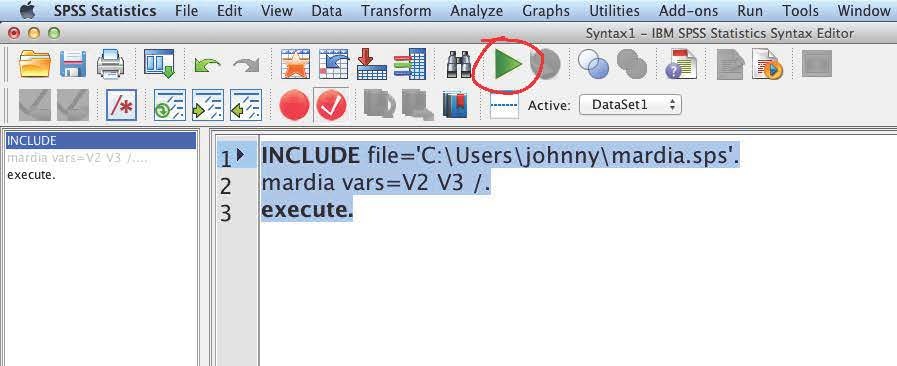
INCLUDE file='C:\Users\johnny\mardia.sps'. mardia vars=V2 V3 /. execute.
Note that you will need to change the folder to the SPSS macro file you just downloaded. Also, the vars to use to calculate the skewness and kurtosis should be changed from V2 V3 to your variables.
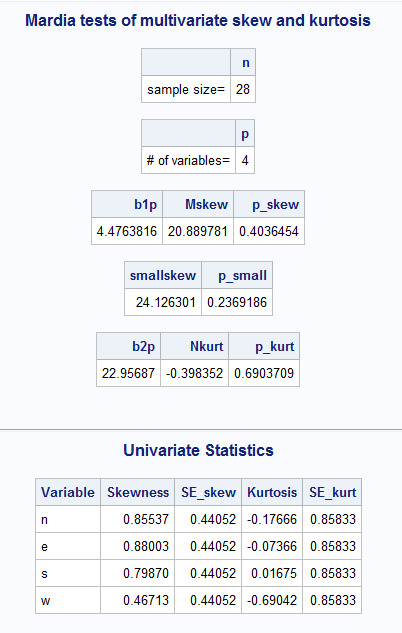
Finally, the output is shown like
SAS
A SAS macro can be used to calculate multivariate skewness and kurtosis. We have edited this macro to get the skewness and kurtosis only.
First, download the macro to your computer, e.g., to C:\Users\johnny\. right click here to download the macro
Second, in the sas script editor, type
%inc "C:\Users\johnny\mardia.sas"; %mardia(data=testdataset, var=V2 V3)
The first line provides the sas macro file location. In the second, one needs to specify the data and variables to use.
An example is shown below
data cork;
input n e s w @@;
datalines;
72 66 76 77 91 79 100 75
60 53 66 63 56 68 47 50
56 57 64 58 79 65 70 61
41 29 36 38 81 80 68 58
32 32 35 36 78 55 67 60
30 35 34 26 46 38 37 38
39 39 31 27 39 35 34 37
42 43 31 25 32 30 30 32
37 40 31 25 60 50 67 54
33 29 27 36 35 37 48 39
32 30 34 28 39 36 39 31
63 45 74 63 50 34 37 40
54 46 60 52 43 37 39 50
47 51 52 43 48 54 57 43
;
%inc "C:\Users\johnny\mardia.sas";
%mardia(data=cork, var=n e s w)
The sample output looks like this
STATA
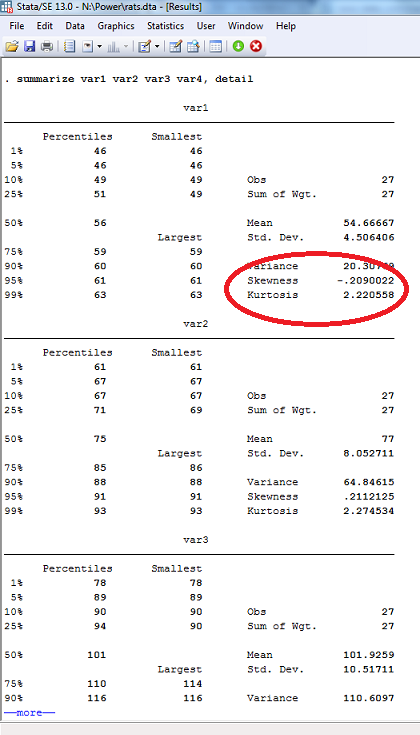
Univariate skewness and kurtosis can be calculated in STATA along with other descriptive statistics by adding detail as an option to the summarize command:
summarize var1 var2 var3 var4, detail
Just change var1, var2, etc. to the variables of interest in your data set. Skewness and kurtosis will appear in the lower right corner of each variable's output
Multivariate skewness and kurtosis can then be calculated using the following syntax:
mvtest normality var1 var2 var3 var4, stats(skewness kurtosis)
Again, specify the appropriate variable names. The sample output will appear as
FAQ
Can you calculate the statistics for me?
- Yes! If you send us the data, we can do the calculation by ourselves and delete the data from our computer once it is done.
Which variables should I use?
- We are interested in the skewness and kurtosis of all the continuous variables (including ordinal ones such as likert scale variables) you have used in the data analysis of your paper. Therefore, do not include categorical, binomial, or proportional data, etc. Note that this means to not include ID, condition #, or other such variables in your calculations. If you have used composite scores such as mean or sum scores, please calculate skewness and kurtosis for them instead of the item scores.
Should I include reverse codings?
- If items have been reverse-coded, please make sure to only include one or the other.
What if I didn't use any continuous variables in my paper?
- If there are no suitable variables from your paper then please ignore our email. Nevertheless, we appreciate your interest in our study!
What if my paper has more than one study?
- Please calculate skewness and kurtosis separately for each study. This is especially important for multivariate skewness and kurtosis as we only want those measures of a group of variables used together in the same analysis.
What if my data is in a format not listed here?
- Convert your data to csv format and use the online calculator. Please let us know if you run into trouble.
What criteria did you use for selecting papers?
- We are interested in the skewness/kurtosis of all papers published in Psychological Science and the American Educational Research Journal in the past 5 years.